


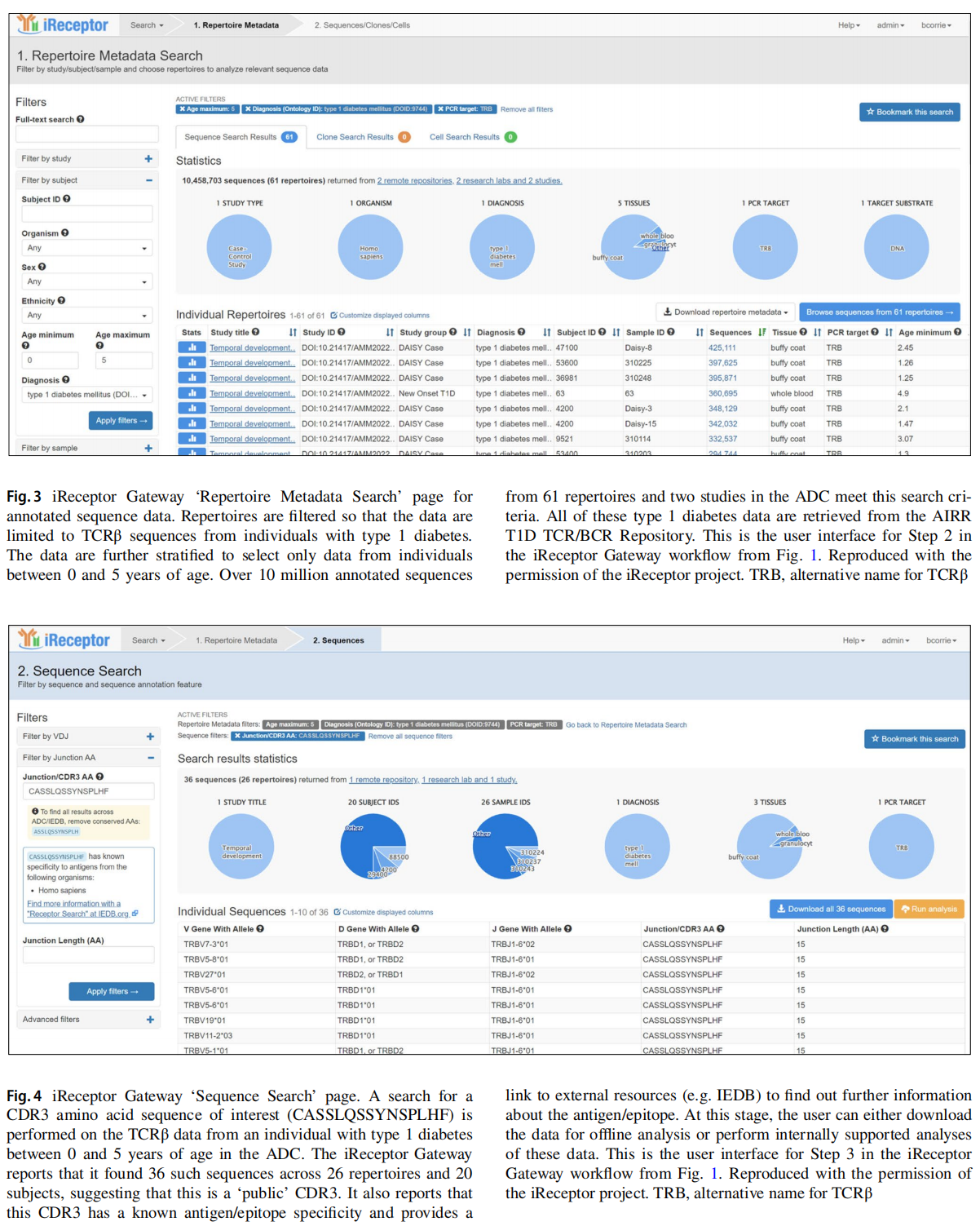
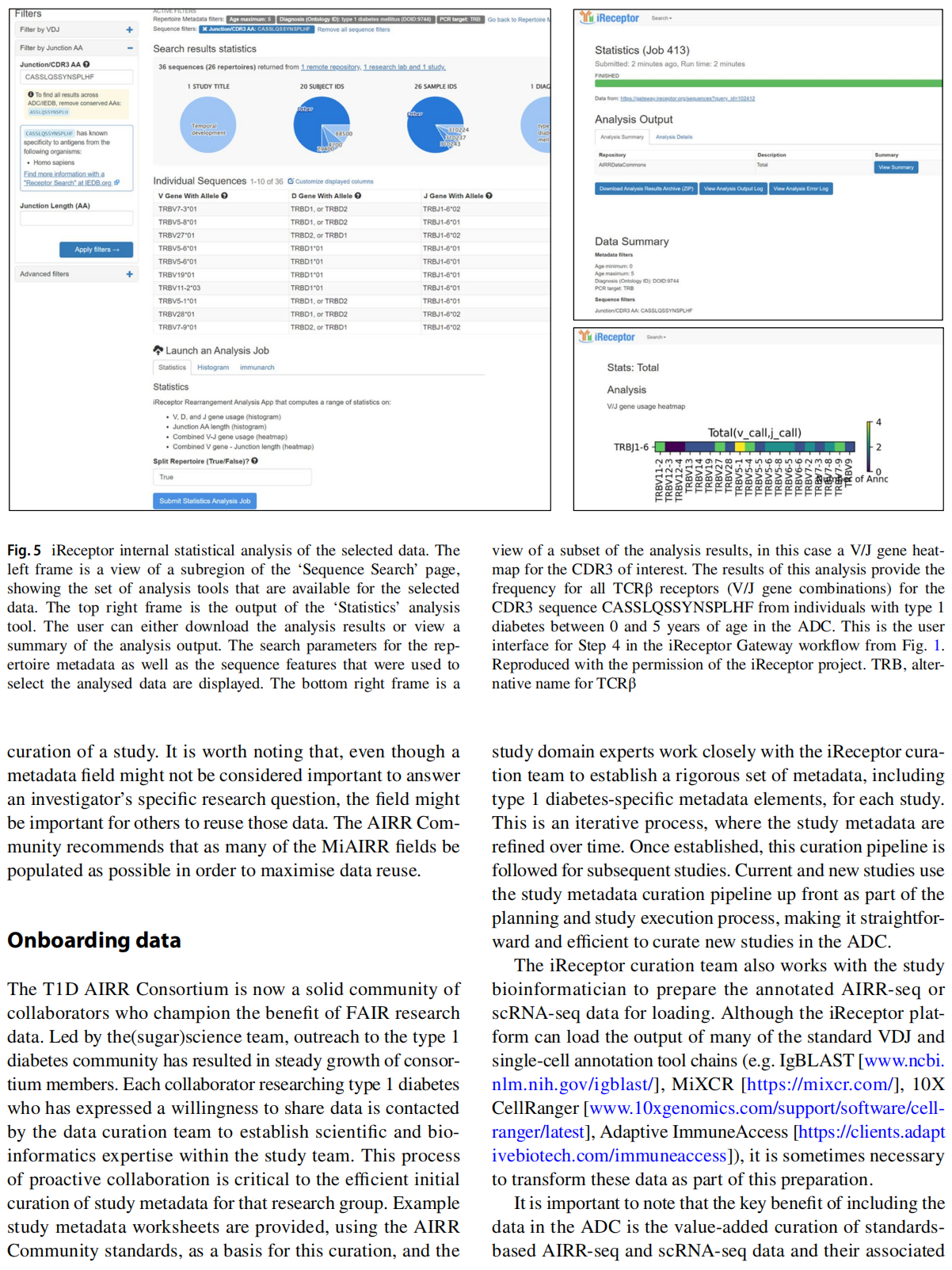

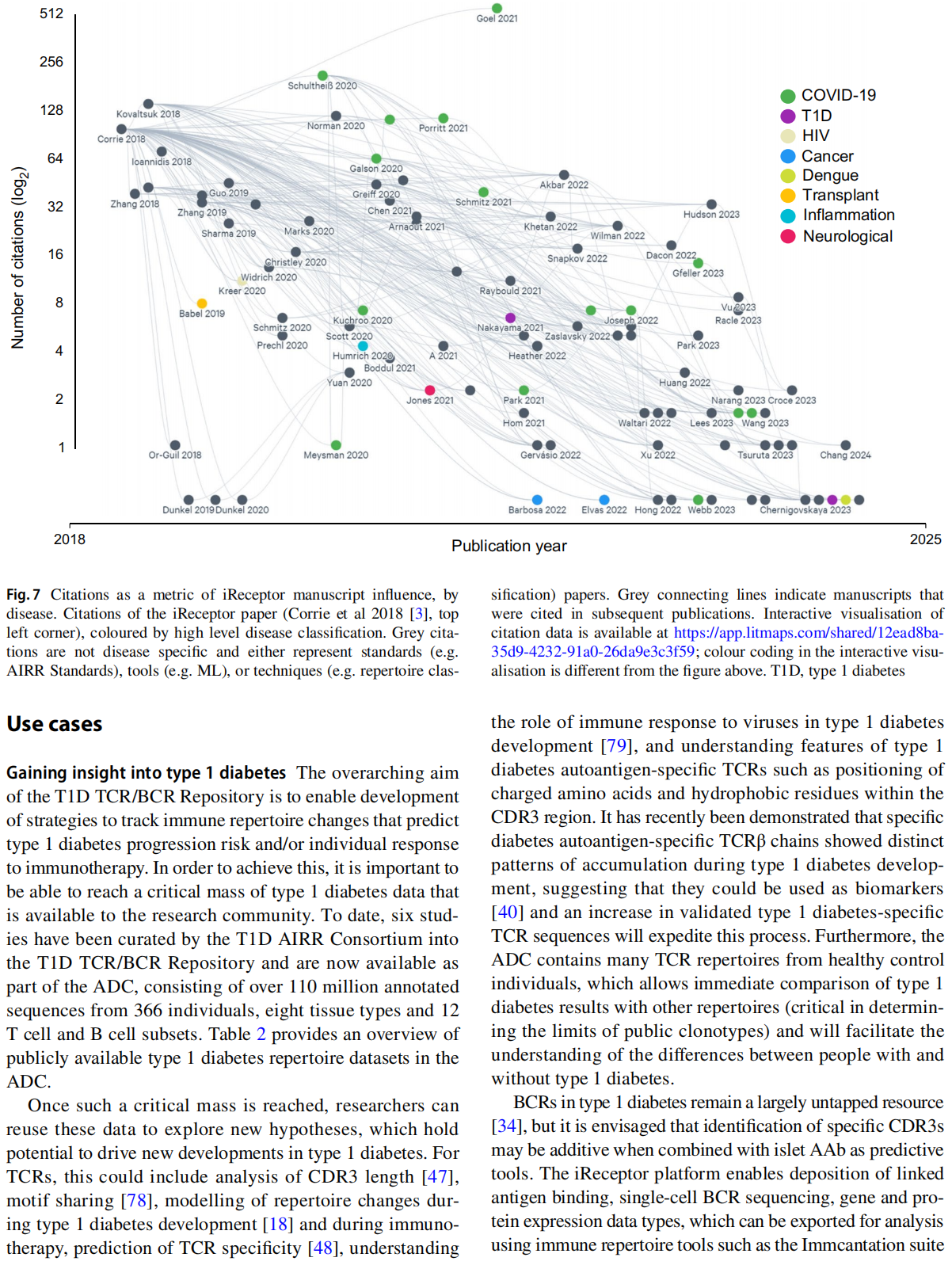
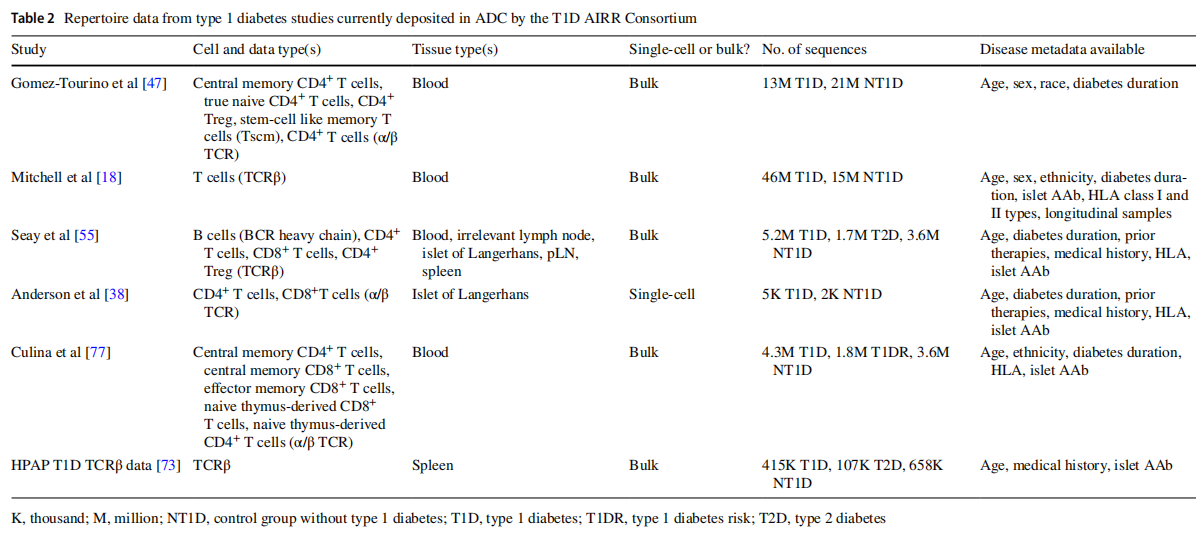





This article is excerpted from the Diabetologia (2025) 68:186-202 by Wound World.
Stephanie J. Hanna1 · Rachel H. Bonami2,3,4,5 · Brian Corrie6,7 · Monica Westley8 · Amanda L. Posgai9 · Eline T. Luning Prak10 · Felix Breden6,7 · Aaron W. Michels11 · Todd M. Brusko9,12,13 · Type 1 Diabetes AIRR Consortium
Received: 26 May 2024 / Accepted: 19 August 2024 / Published online: 29 October 2024
© The Author(s) 2024
Extended author information available on the last page of the article
Abstract
Human molecular genetics has brought incredible insights into the variants that confer risk for the development of tissuespecific autoimmune diseases, including type 1 diabetes. The hallmark cell-mediated immune destruction that is characteristic of type 1 diabetes is closely linked with risk conferred by the HLA class II gene locus, in combination with a broad array of additional candidate genes influencing islet-resident beta cells within the pancreas, as well as function, phenotype and trafficking of immune cells to tissues. In addition to the well-studied germline SNP variants, there are critical contributions conferred by T cell receptor (TCR) and B cell receptor (BCR) genes that undergo somatic recombination to yield the Adaptive Immune Receptor Repertoire (AIRR) responsible for autoimmunity in type 1 diabetes. We therefore created the T1D TCR/ BCR Repository (The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository) to study these highly variable and dynamic gene rearrangements. In addition to processed TCR and BCR sequences, the T1D TCR/BCR Repository includes detailed metadata (e.g. participant demographics, disease-associated parameters and tissue type). We introduce the Type 1 Diabetes AIRR Consortium goals and outline methods to use and deposit data to this comprehensive repository. Our ultimate goal is to facilitate research community access to rich, carefully annotated immune AIRR datasets to enable new scientific inquiry and insight into the natural history and pathogenesis of type 1 diabetes.
Keywords AIRR · AIRR Data Commons · Autoantibodies · B cell receptors · FAIR data · Next-generation sequencing · Single-cell RNA-seq · T cell receptors · Type 1 diabetes
Abbreviations
AAb Autoantibody/autoantibodies
ADC AIRR Data Commons
AIM Activation-induced marker
AIRR Adaptive Immune Receptor Repertoire
AIRR-seq AIRR sequencing
BCR B cell receptor
CDR3 Complementarity determining region 3
FAIR Findable, Accessible, Interoperable, Reusable
GEO Gene Expression Omnibus
HPAP Human Pancreas Analysis Program
IEDB Immune Epitope Database
MiAIRR Minimal information about AIRR
ML Machine learning
pLN Pancreatic lymph node(s)
SARS-CoV-2 Severe acute respiratory syndrome coronavirus 2
scRNA-seq Single-cell RNA-seq
SRA Sequence Read Archive
T1D TCR/BCR Repository The Type 1 Diabetes T Cell Receptor and B Cell Receptor Repository
TCR T cell receptor
TCRβ T cell receptor β chain
Tfh T follicular helper
Treg Regulatory T cell(s)
VDJ Variable, diversity and joining gene segments
Stephanie J. Hanna and Rachel H. Bonami contributed equally to this work. Aaron W. Michels and Todd M. Brusko contributed equally as joint senior authors.
Members of the Type 1 Diabetes AIRR Consortium are listed in the Acknowledgements.

















This article is excerpted from the Diabetologia (2025) 68:186-202 by Wound World.
