
伤口世界
电子邮件地址: 该Email地址已收到反垃圾邮件插件保护。要显示它您需要在浏览器中启用JavaScript。

- 星期六, 28 2月 2026
Diabetes duration and types of diabetes treatment in data-driven clusters of patients with diabetes


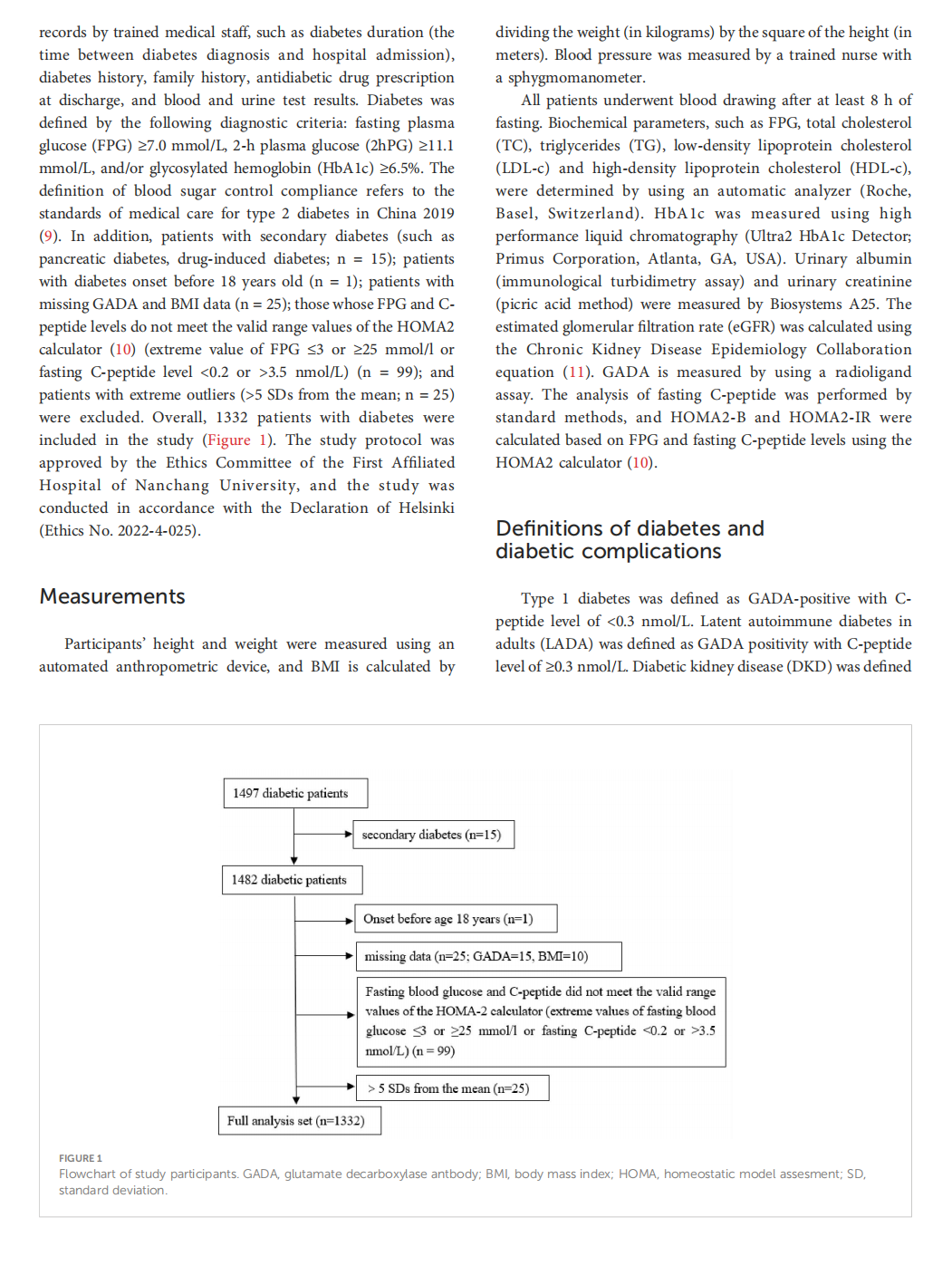

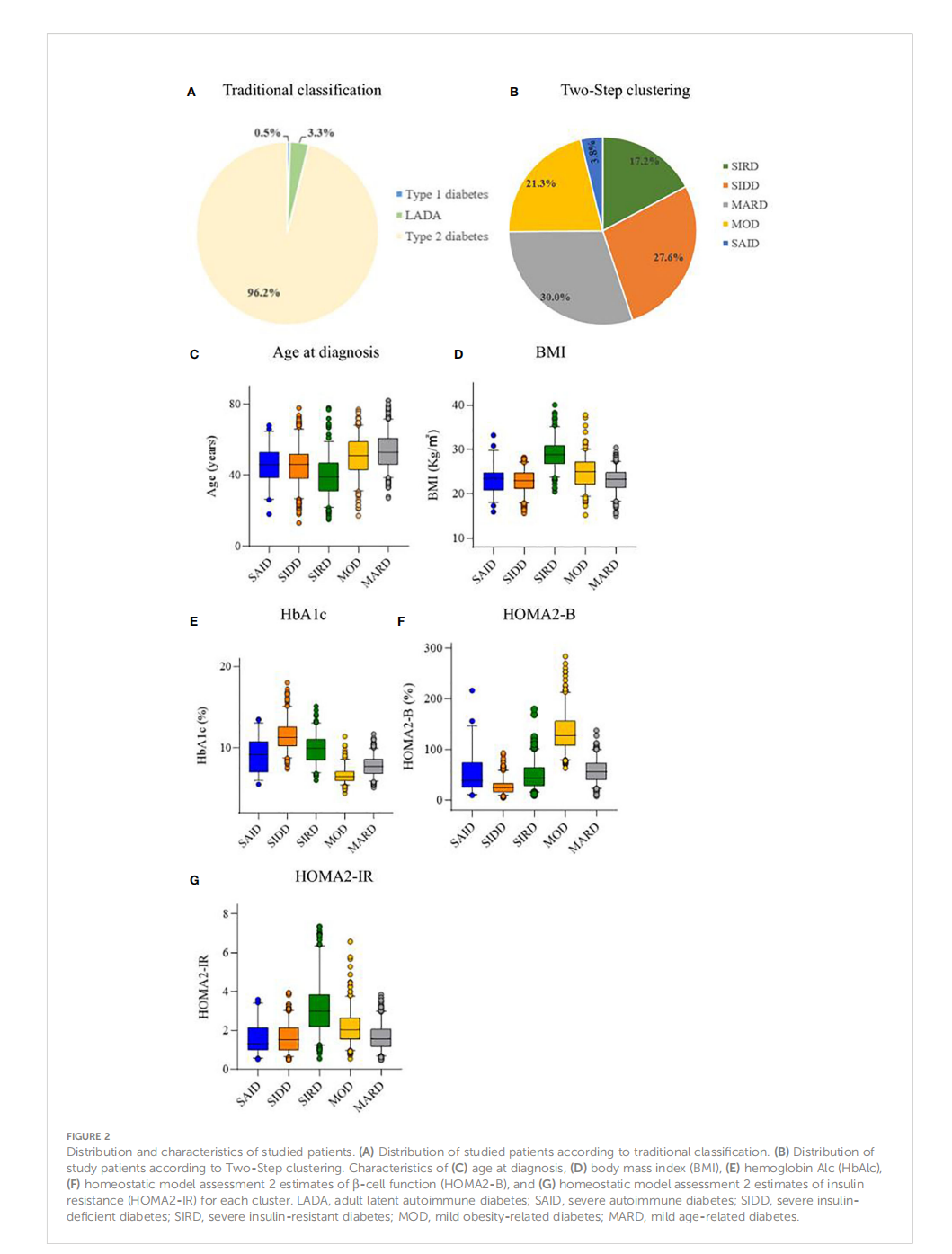
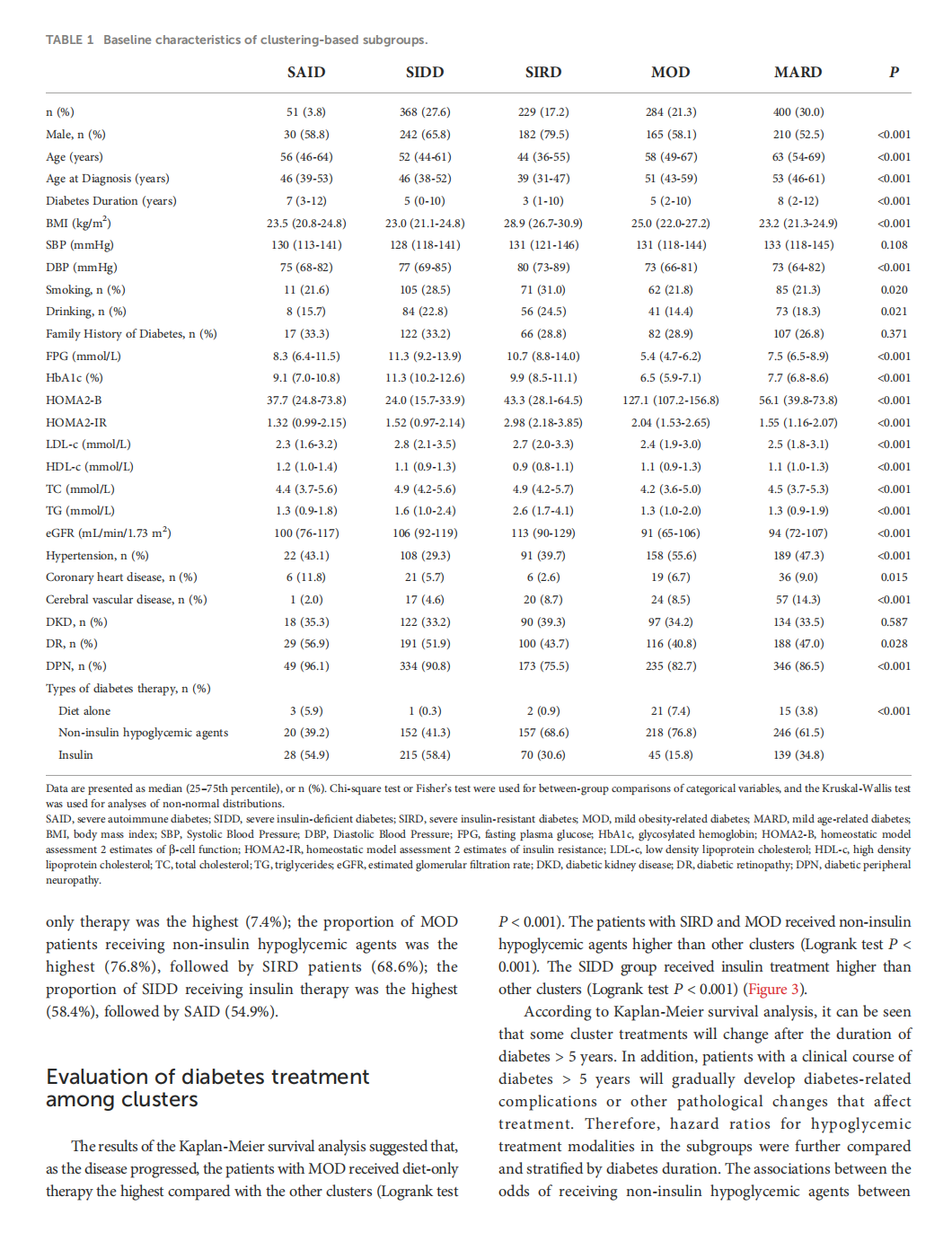
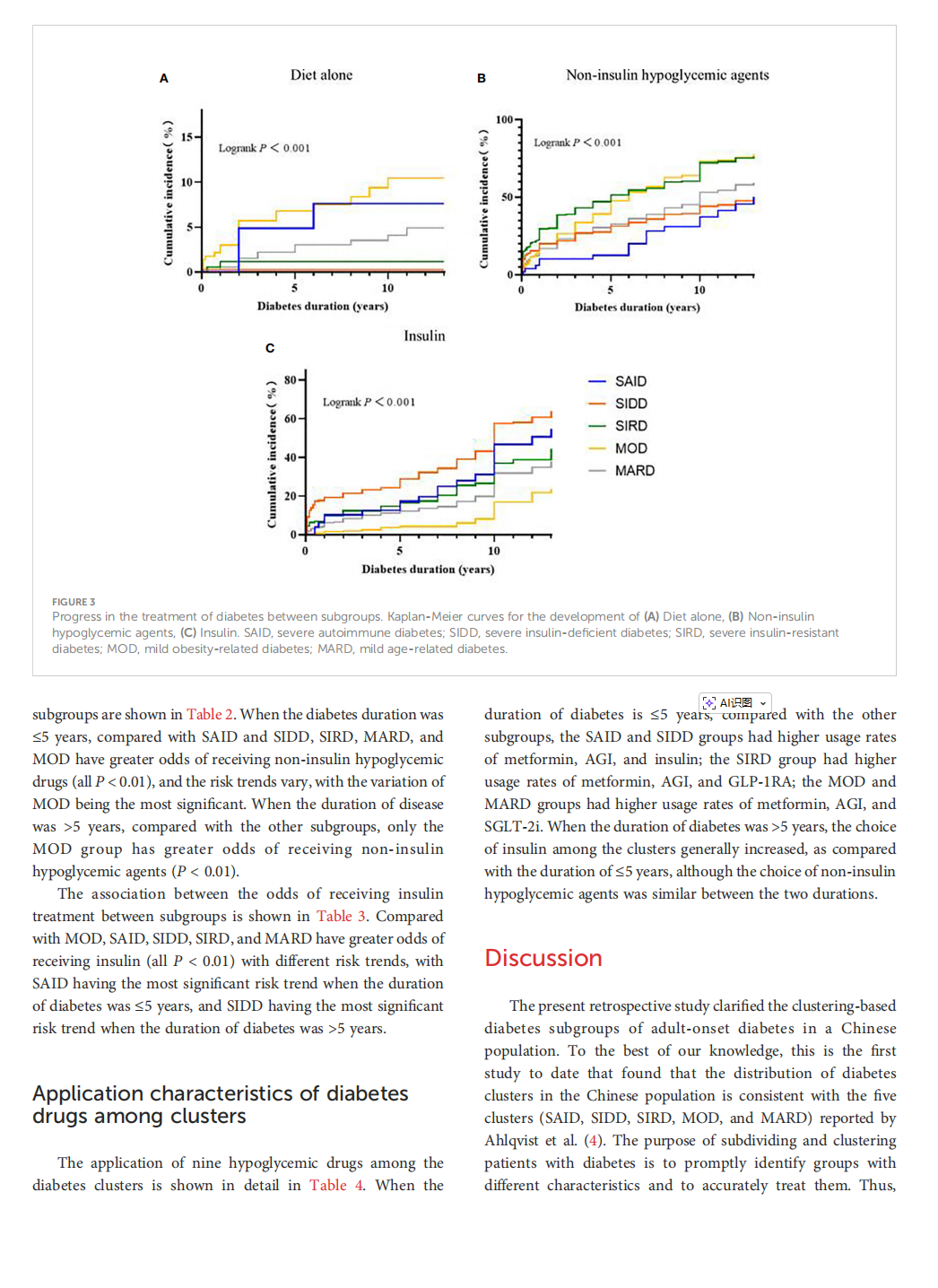
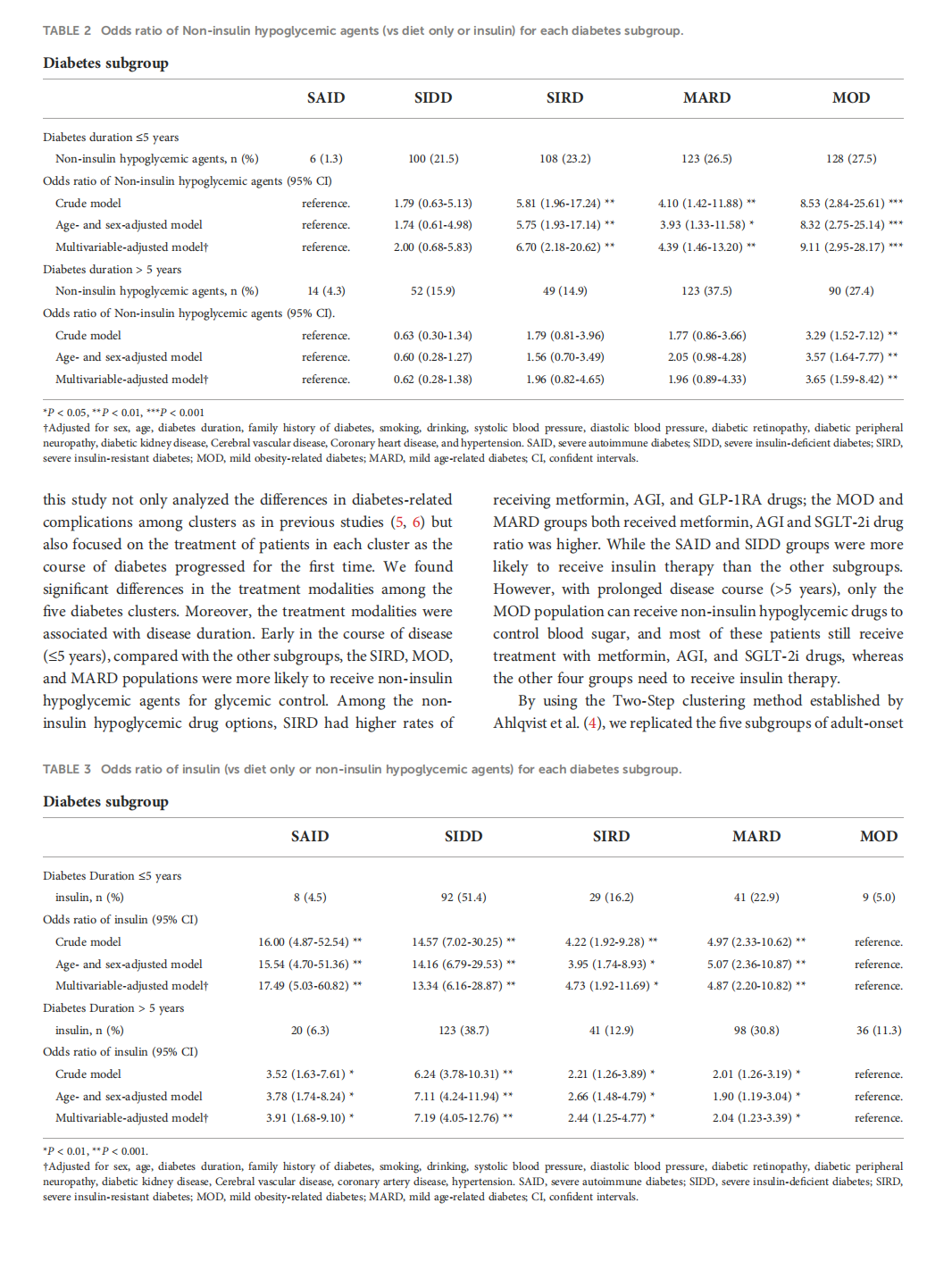
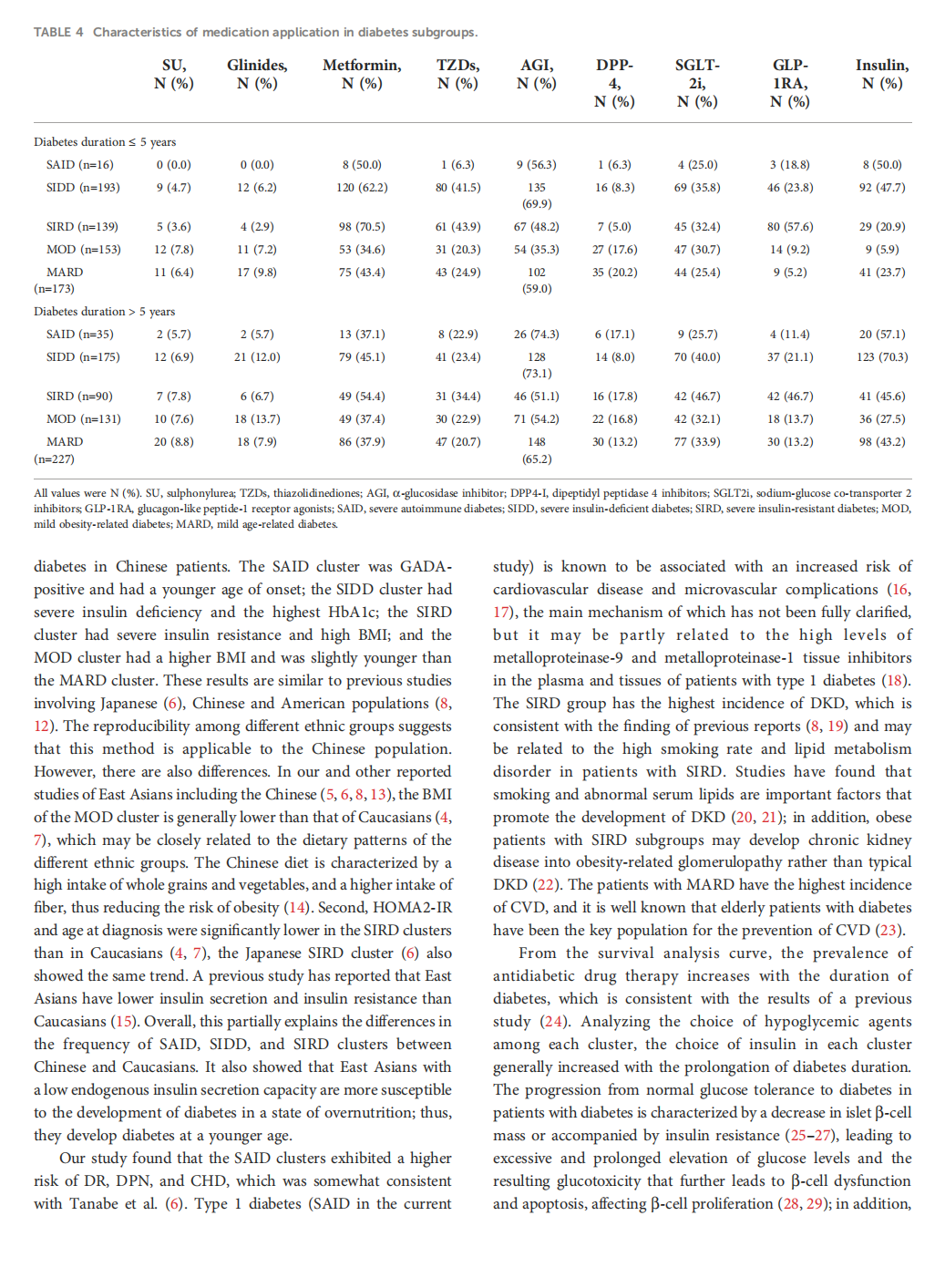



This article is excerpted from the 《 Frontiers in Endocrinology》 by Wound World

- 星期五, 27 2月 2026
The mediating role of diabetes stigma and self-efficacy in relieving diabetes distress among patients with type 2 diabetes mellitus: a multicenter cross-sectional study

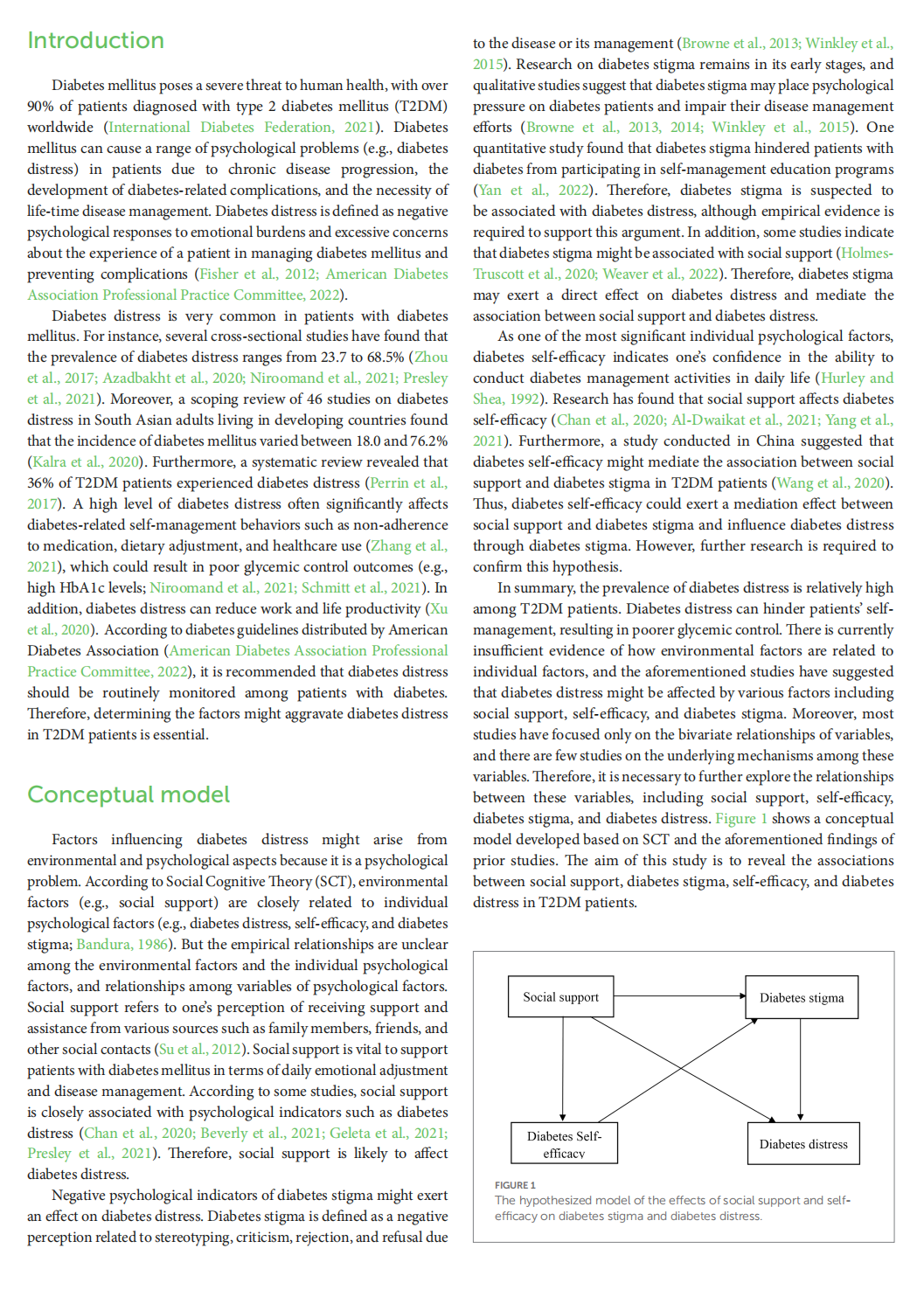


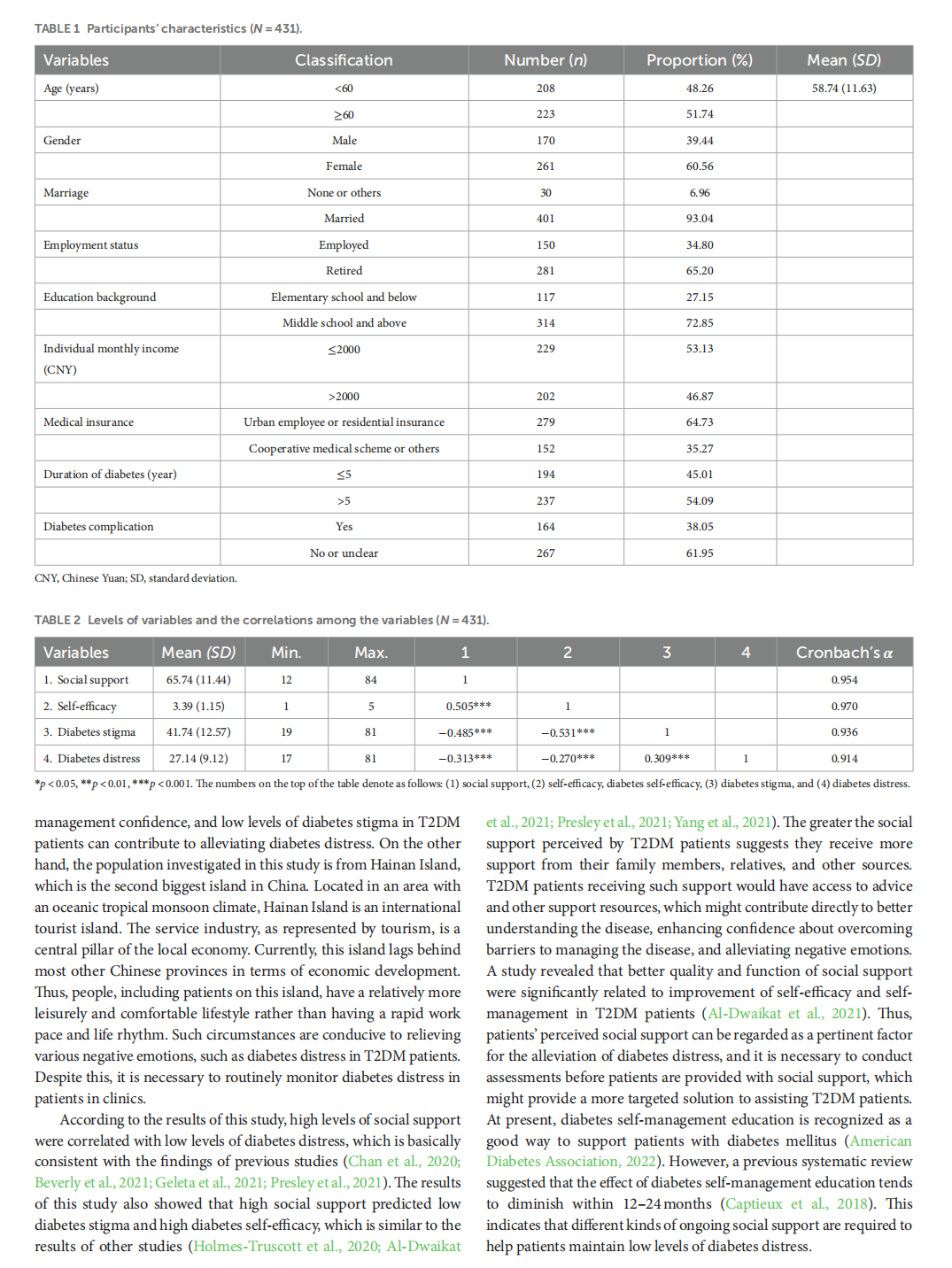
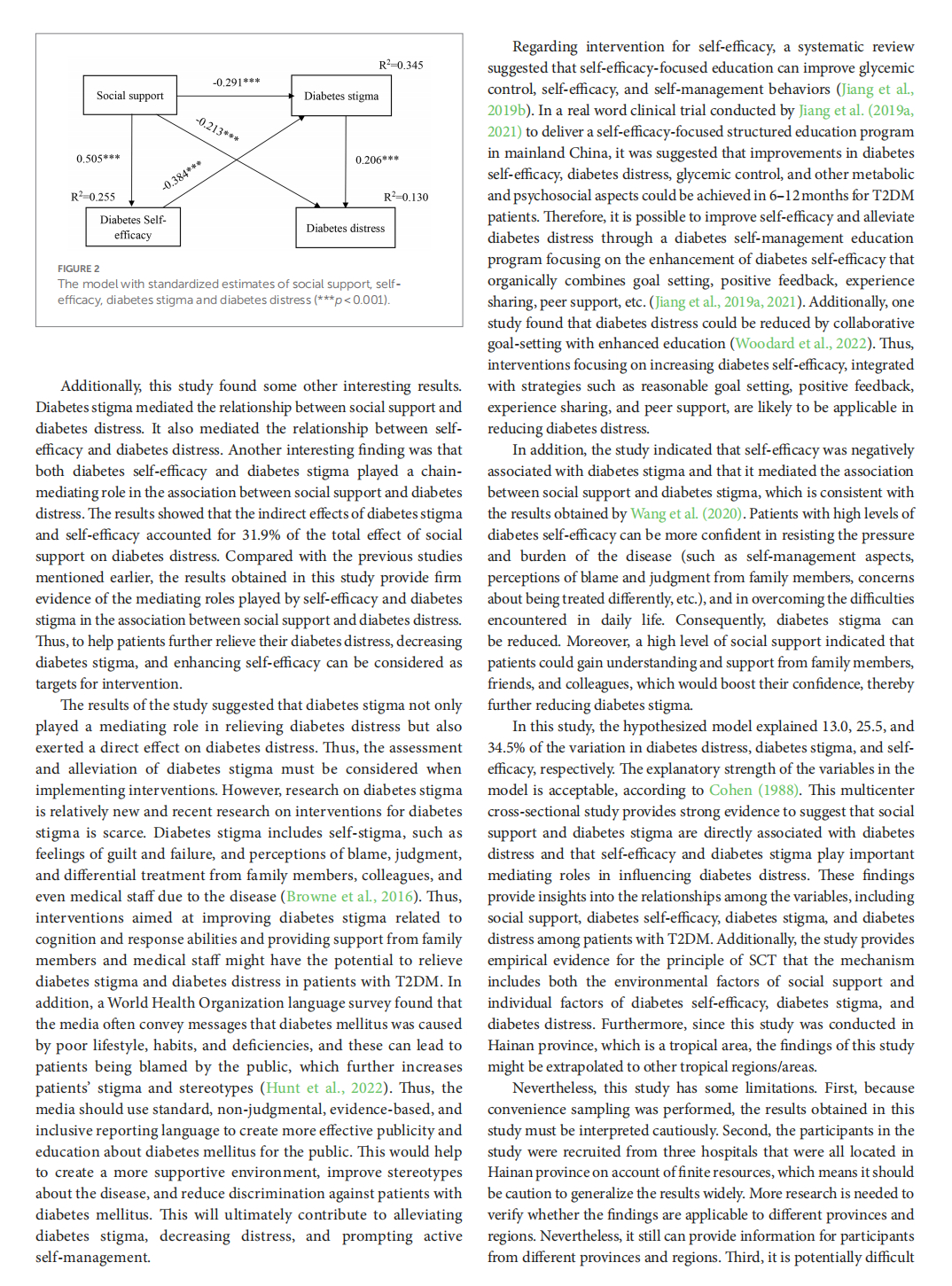


This article is excerpted from the 《Frontiers in Psychology》 by Wound World


- 星期六, 14 2月 2026
Deep learning techniques for imaging diagnosis of renal cell carcinoma: current and emerging trends

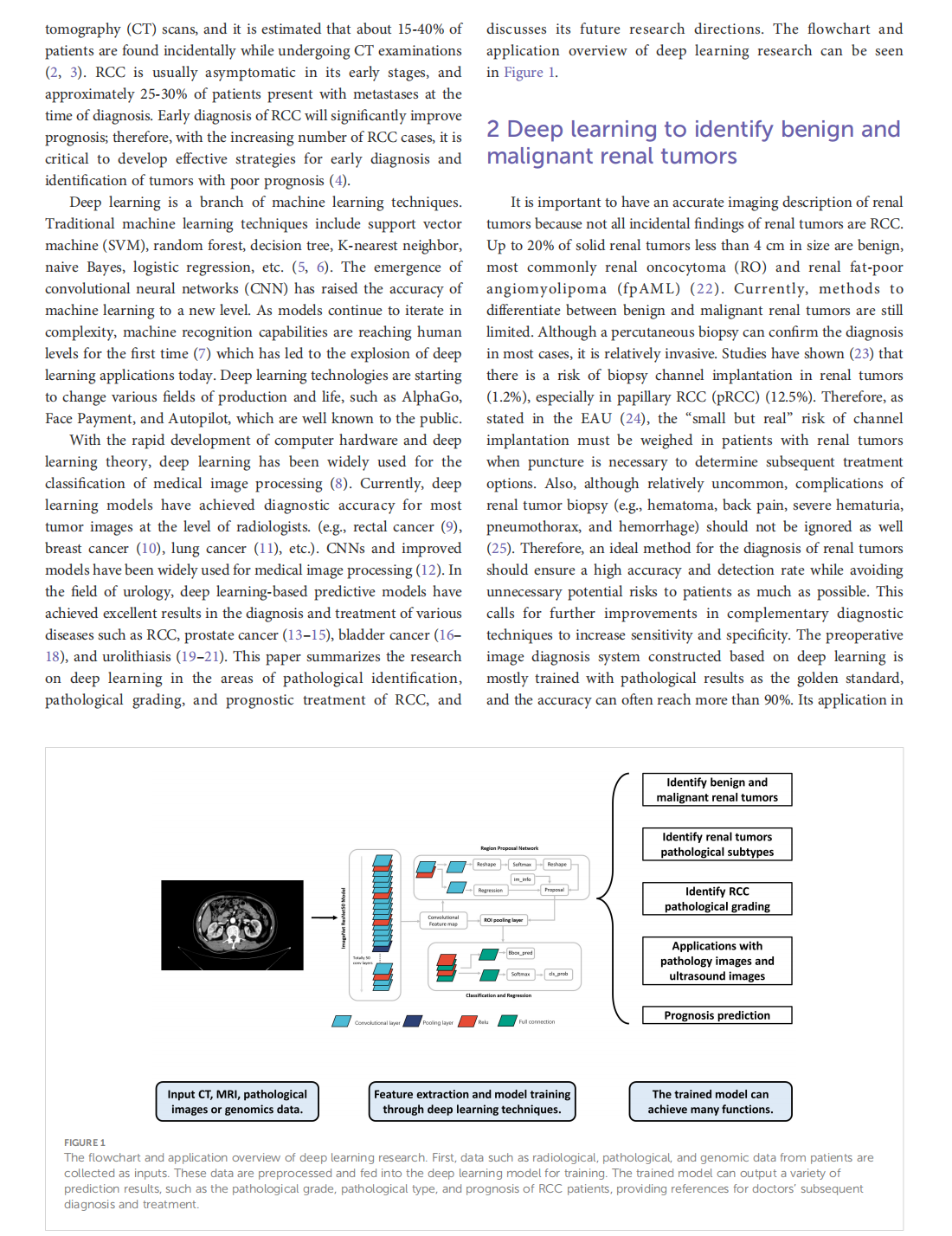
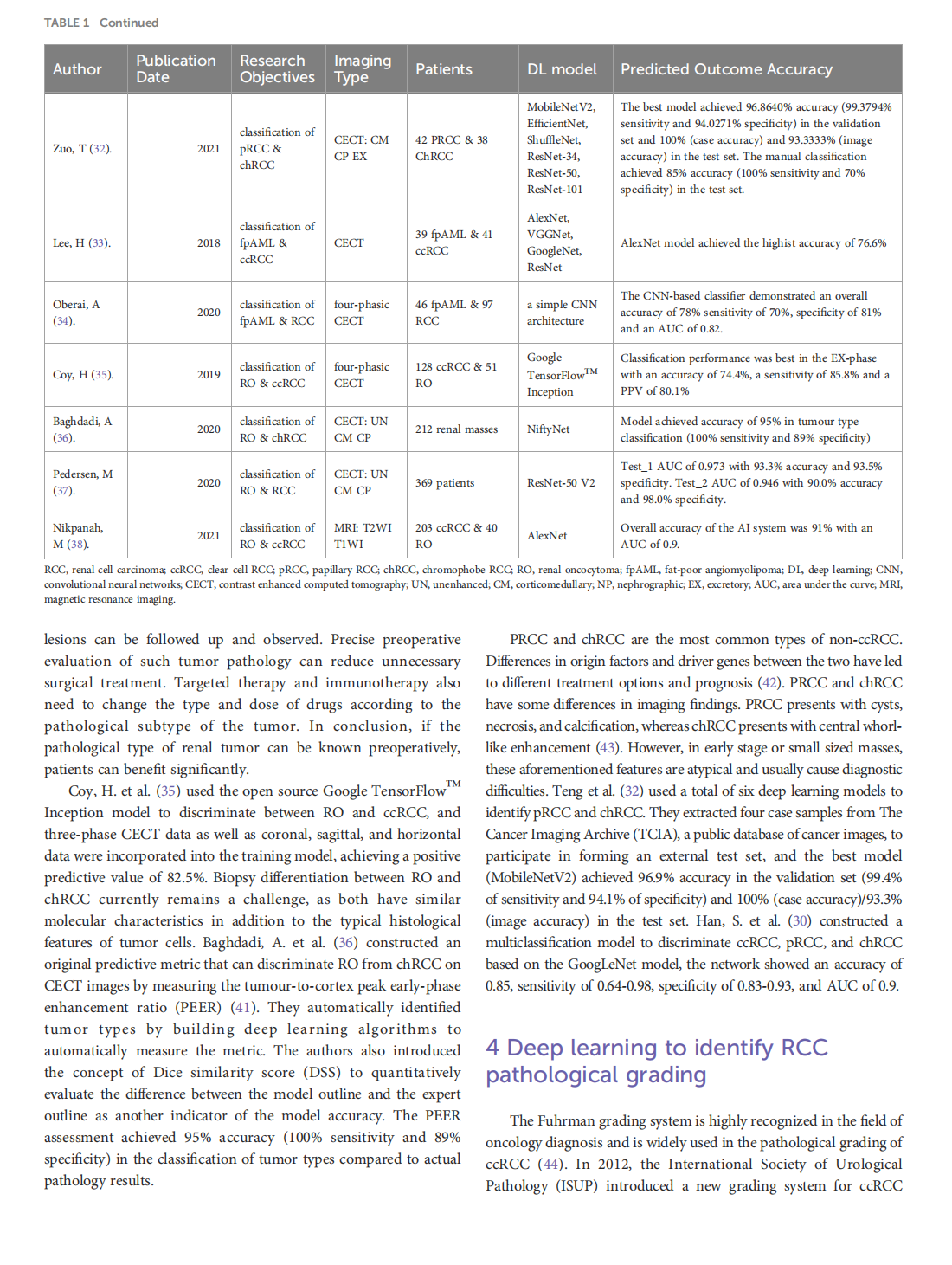
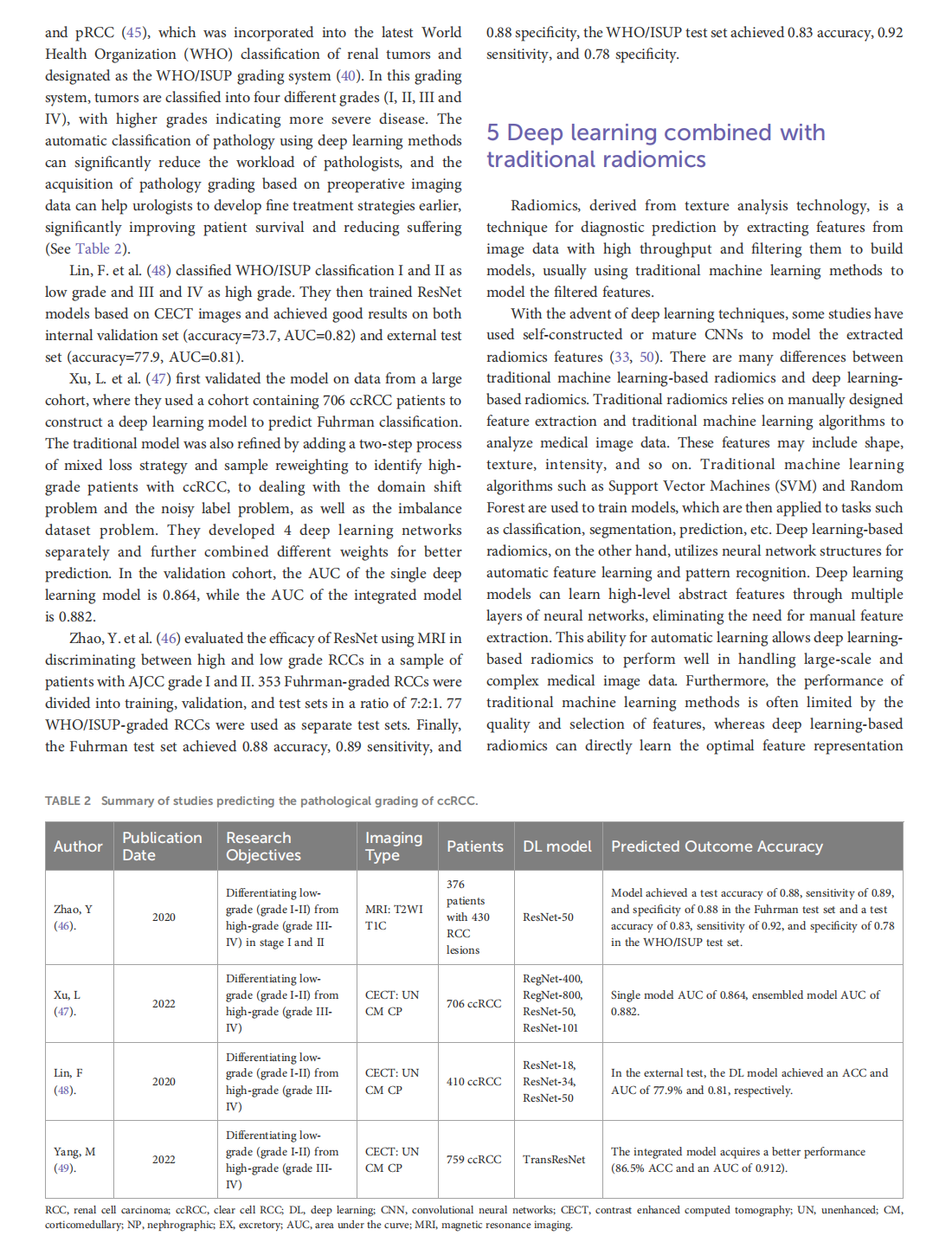
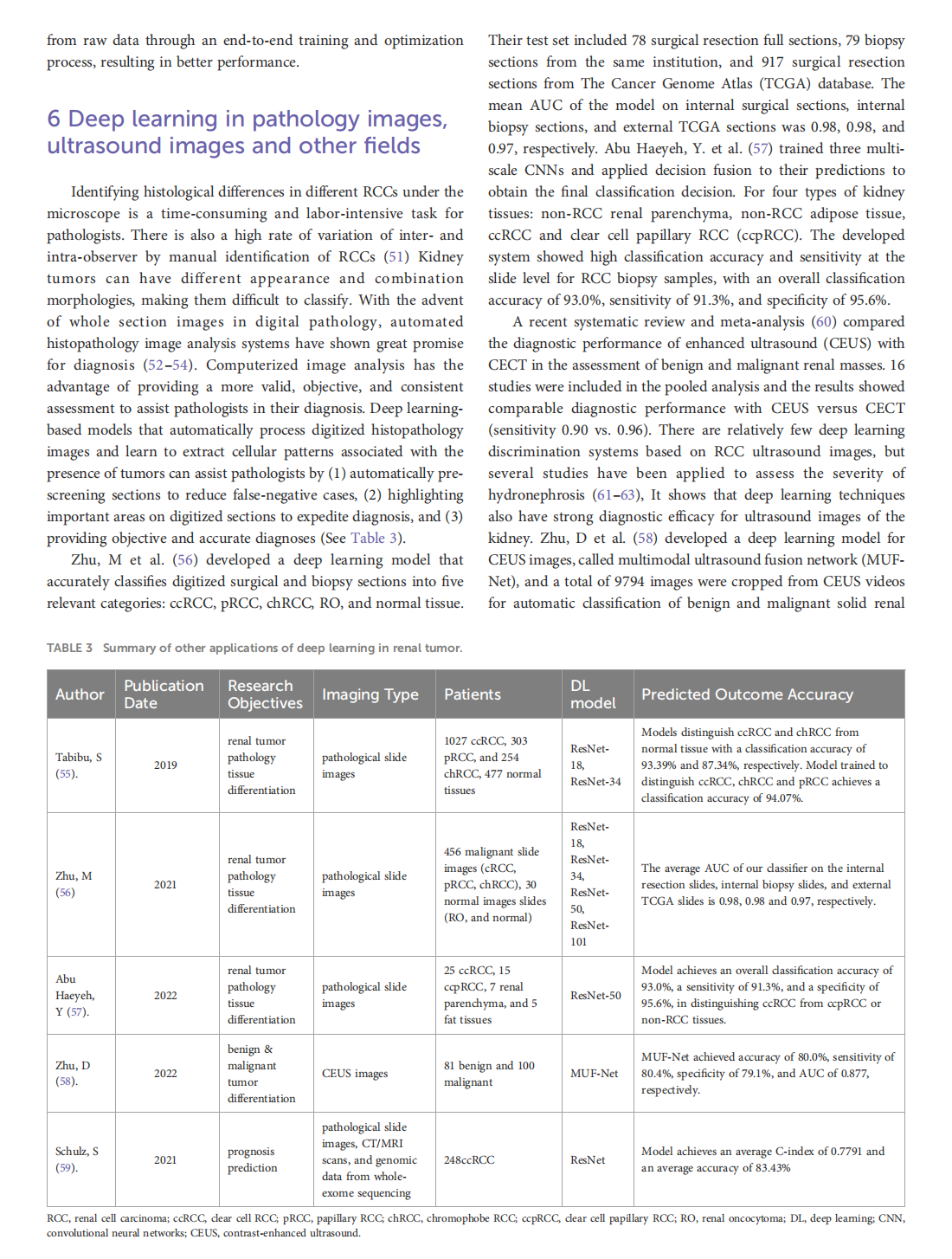





This article is excerpted from the《Frontiers in Oncology》by Wound World

- 星期三, 11 2月 2026
Medical treatment of acquired uterine arteriovenous fistulae related to pregnancy: 2 case reports and literature review

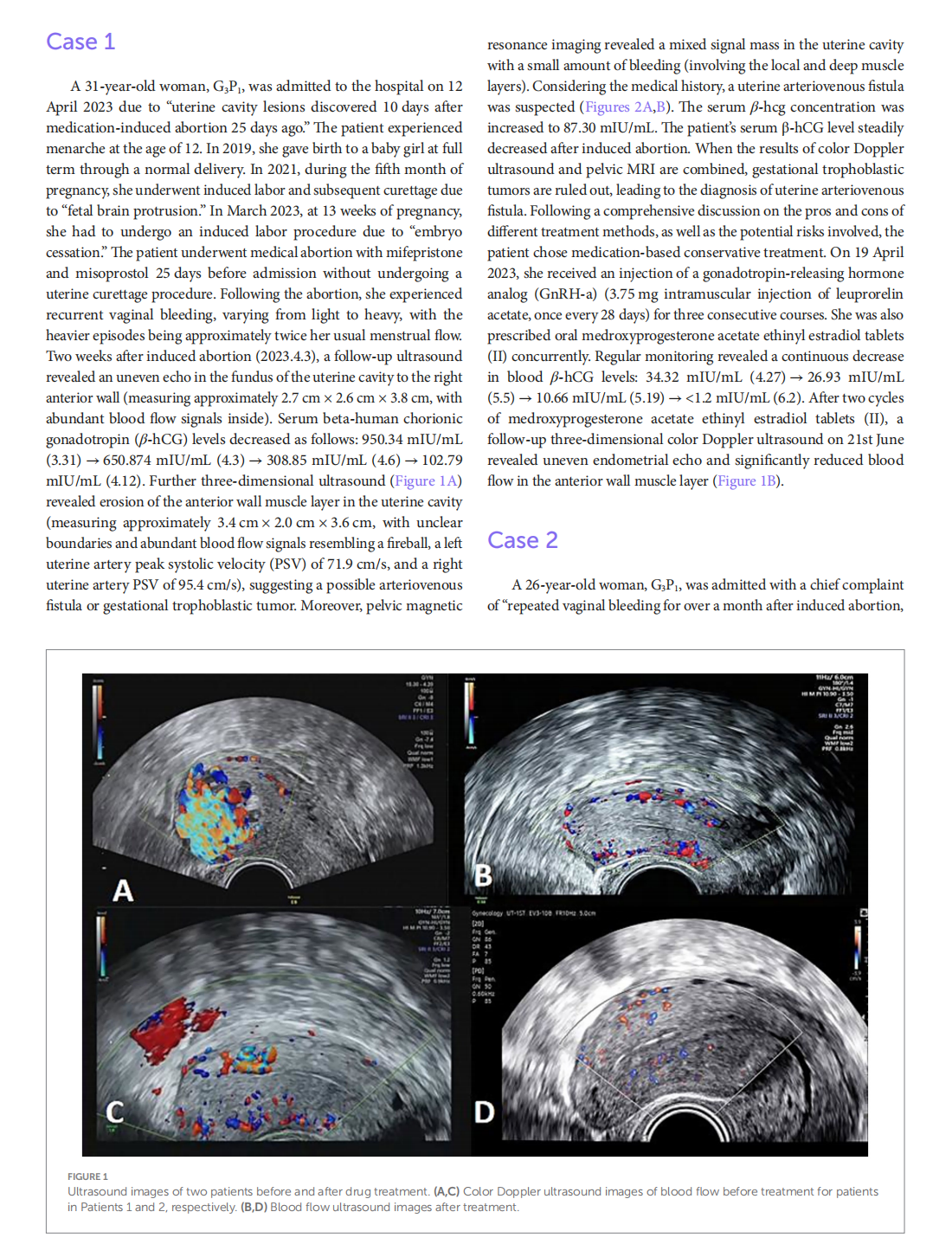
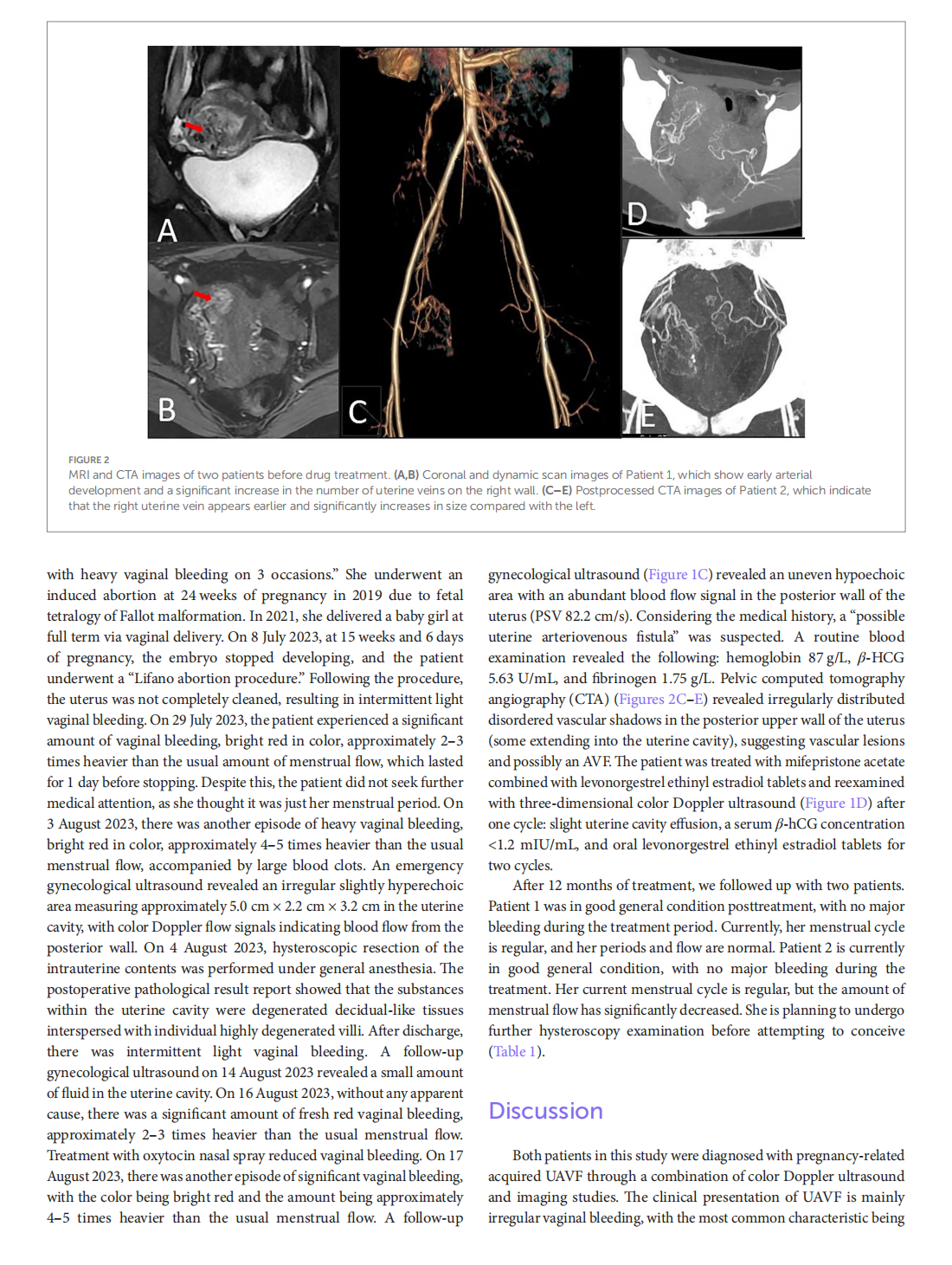
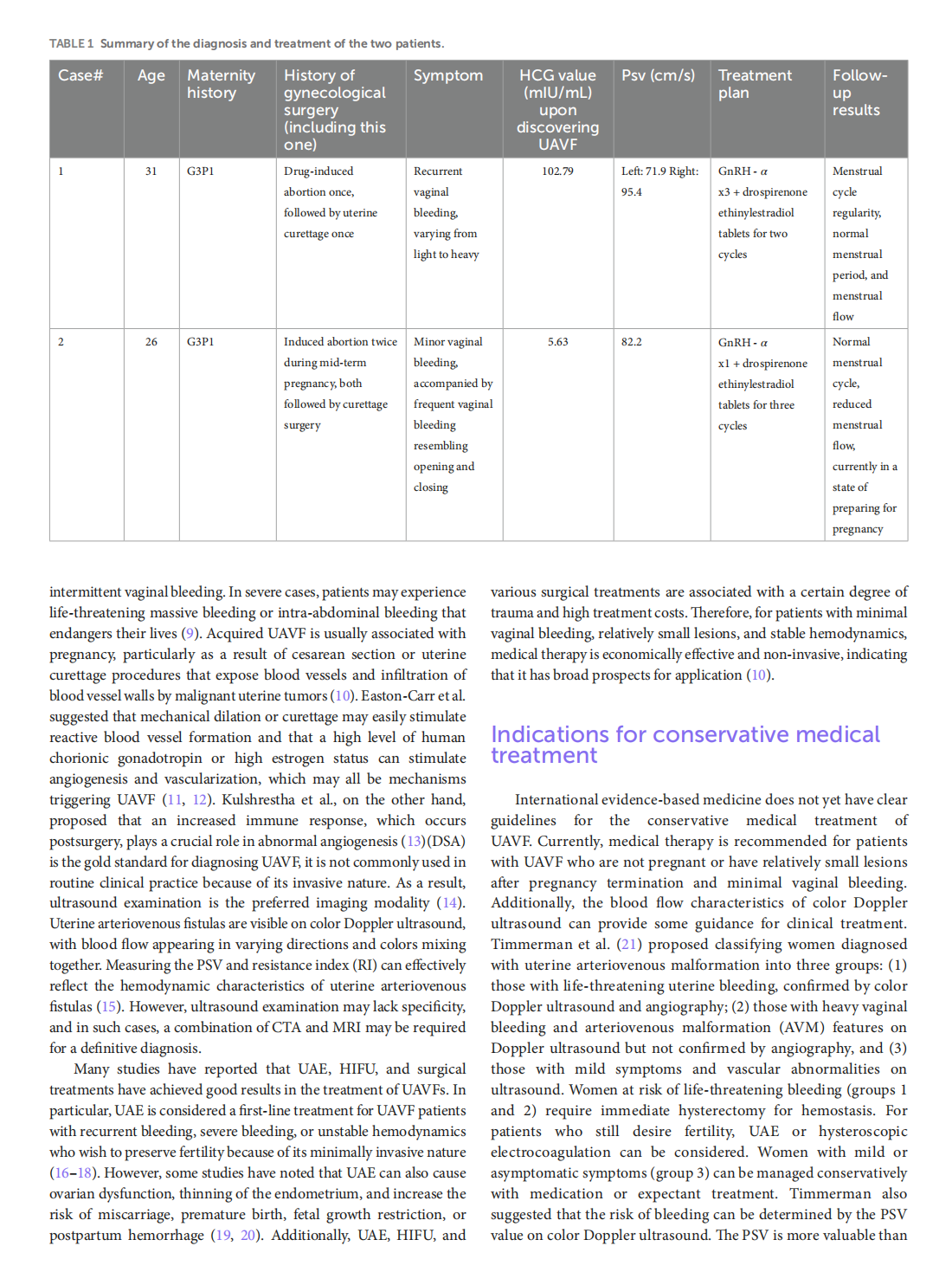



This article is excerpted from the 《Frontiers in Medicine》 by Wound World

- 星期一, 09 2月 2026
Virtual reality as a non-medical tool in the treatment of anxiety, pain, and perception of time in children in the maintenance phase of acute lymphoblastic leukemia treatment



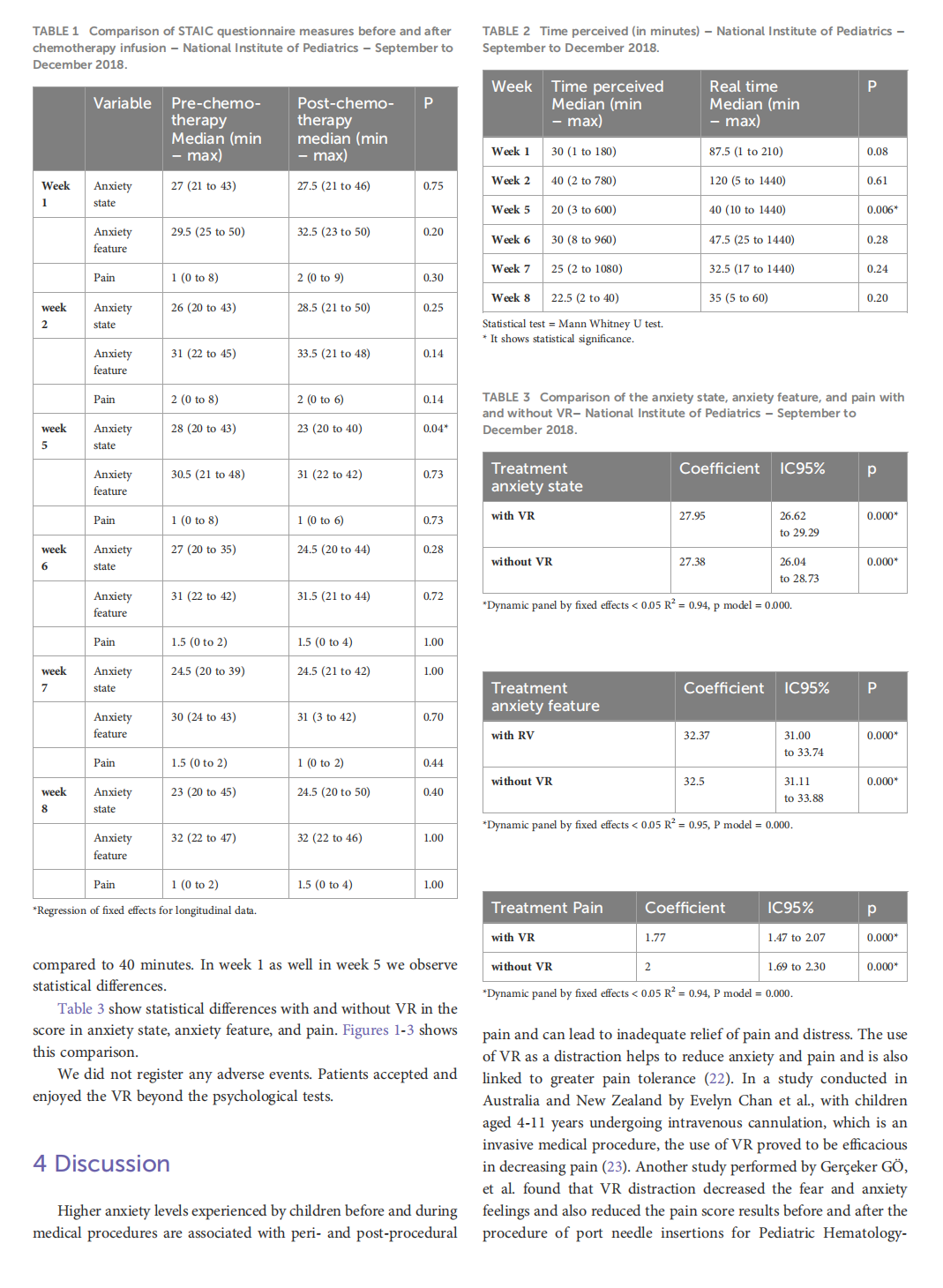
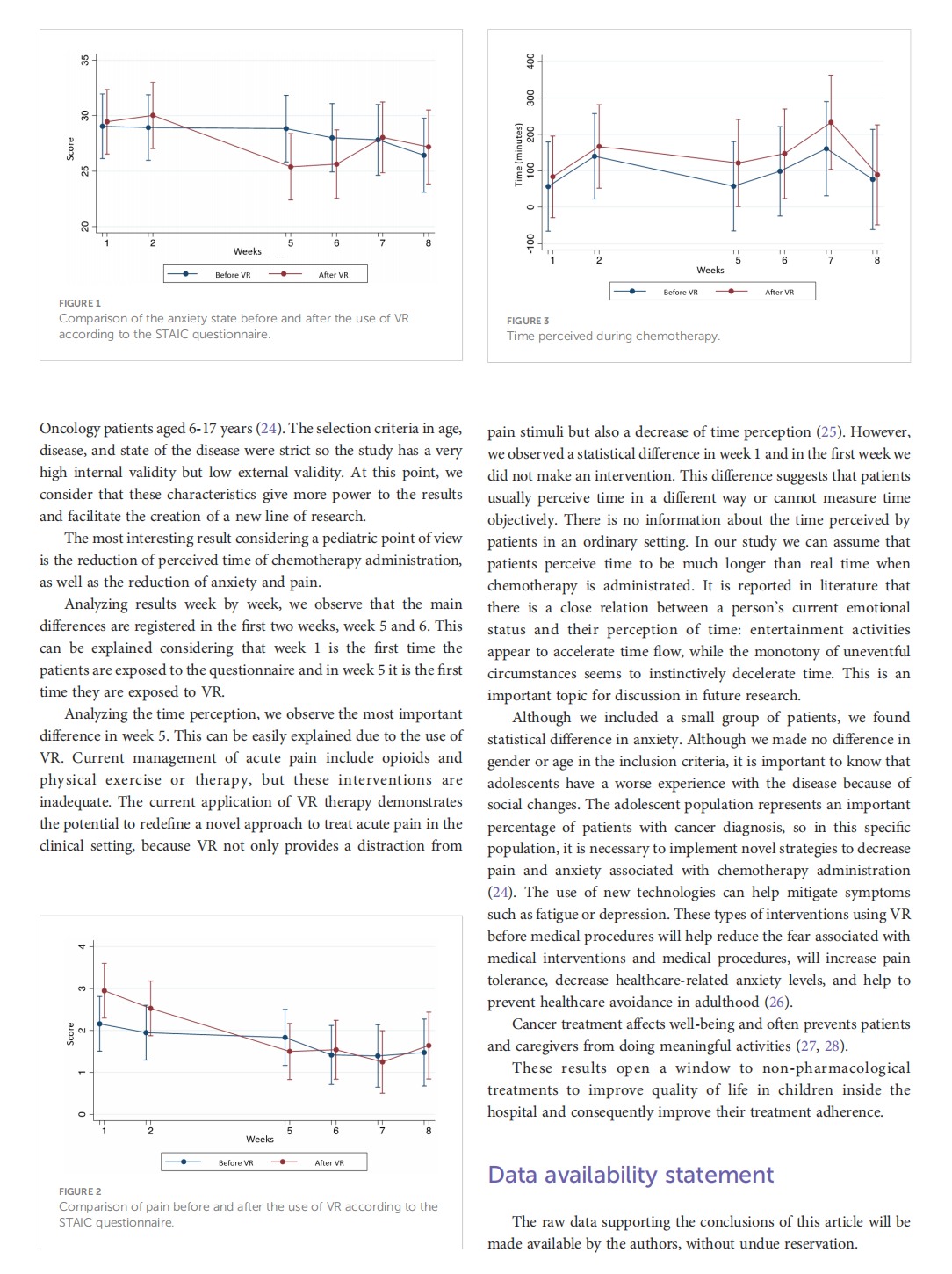


This article is excerpted from the《Frontiers in Oncology》by Wound World


